how to find dropout rate in 10x genomics data
Swimming in a body of water of cells: Why unmarried-cell genomics?
Complex biological systems issue from the coordinated part of individual cells performing an intricate "dance," each contributing their own special role to the ensemble. Because of this complexity, gene expression research in organisms, tissues or cell populations is often limited past using traditional bulk RNA-seq methods. Every bit scientists, nosotros intuitively sympathise that when we "grind and find" in social club to perform experiments on our diluted mix of RNA, we are really only getting a minor thought of what might be going on in our cells of interest: cell-to-cell differences are missed and cellular heterogeneity can exist completely masked. In fact, bulk RNA expression analysis ofttimes describes an inferred land in which none (or very few) of the cells actually be!
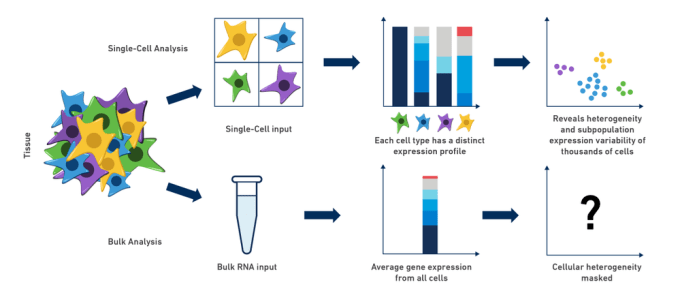
Possibly y'all are a neuroscientist, working in the mouse brain, studying the development of a sub-population of neurons that produce a specific neurotransmitter. If merely you could isolate that population and analyze it on a jail cell-by-cell basis, the potential insights would be, indeed, neuronally stimulating! Similarly in the instance of cancer research: how do you all-time tease apart gene expression and, perhaps more importantly, cellular heterogeneity, in a tumor? What if you could compare not only inter-tumor, simply intra-tumor differences and explore the immune cell populations related to the tumor microenvironment? This diversity, often obscured by majority RNA-seq methods, can be revealed past using single cell RNA-seq to specifically explore these clinically significant cell populations. The examples of drawbacks to bulk analysis are abundant, and dissecting cell-type differences in nigh, if not all, of these circuitous biological systems is disquisitional to our understanding of their contributions, peculiarly during evolution and in the progression of disease.
How do we isolate and analyze single cells?
10x Genomics' single-cell RNA-seq (scRNA-seq) technology, the Chromium™ Single Cell three' Solution, allows you to analyze transcriptomes on a cell-by-cell basis through the use of microfluidic partitioning to capture single cells and prepare barcoded, side by side-generation sequencing (NGS) cDNA libraries. Specifically, single cells, reverse transcription (RT) reagents, Gel Beads containing barcoded oligonucleotides, and oil are combined on a microfluidic fleck to form reaction vesicles called Gel Chaplet in Emulsion, or GEMs. GEMs are formed in parallel within the microfluidic channels of the chip, allowing the user to procedure 100's to x,000's of single cells in a single 7-minute Chromium™ Instrument run. Information technology's of import to note that cells are loaded at a limiting dilution in order to maximize the number of GEMs containing a single cell to ensure a low doublet charge per unit, while maintaining a loftier cell recovery rate of up to ~65%.
Each functional GEM contains a single jail cell, a single Gel Bead, and RT reagents. Inside each GEM reaction vesicle, a single cell is lysed, the Gel Bead is dissolved to free the identically barcoded RT oligonucleotides into solution, and reverse transcription of polyadenylated mRNA occurs. As a event, all cDNAs from a single cell will have the same barcode, allowing the sequencing reads to exist mapped back to their original unmarried cells of origin. The training of NGS libraries from these barcoded cDNAs is and then carried out in a highly efficient bulk reaction. The video beneath gives a smashing visual explanation of how this all works.
From sample prep to how-to videos: Helpful links for getting started
The Chromium Single Prison cell 3' workflow begins with your cells of involvement followed by NGS library construction using our reagent kits and the Chromium Controller. Preparing your barcoded cDNA library (as by and large described in the higher up video) is an important step and truthful to the saying "garbage in, garbage out" — preparing high quality cell suspensions is key to getting the near out or your RNA-seq data. If you need assist with the cell preparation procedure, we have multiple demonstrated protocols bachelor on our website such every bit dissociating neural tissue or preparing moss protoplast suspensions for use in scRNA-seq, with more than protocols being added regularly on our support website. For guidance and recommendations on how to best prep your samples once they are in suspension, a good starting point is our single cell preparation guide, where you lot can learn more about best practices and view a general protocol. If you are more of a visual learner, our how-to-videos, especially Capacity four through 7, should help walk y'all through the sample and library grooming procedure.



-
Unmarried Cell Prep Guide
-
Demonstrated Protocols
-
How-to-videos
-
Chromium™ Controller
-
User Guides
-
How-to-videos
-
Cell Ranger™ Software
-
Loupe™ Jail cell Browser
-
How-to-videos
Analyzing your data, 1 cell at a time
After your library is prepared, sequencing is performed using Illumina® sequencing instruments. This leads to the fun part: gleaning biological insights from your unmarried cell RNA-seq information! The (potentially) massive amount of information you generate from the Chromium Single Jail cell 3' Solution can be hands handled past our software and visualization tools that were designed to take away a lot of the guesswork—Jail cell Ranger™ Software transforms the barcoded sequencing data into files ready for single cell expression analysis, and visualization is accomplished via the Loupe™ Cell Browser. If you would like a step-past-footstep tutorial on using our software, Chapters 8 through xi of our how-to-videos cover everything from transcript counting with Prison cell Ranger Software to beautiful data visualization with Loupe Prison cell Browser. There are also a growing number of unmarried cell analysis tools developed past the customs, including Seurat, Monocle and Cell View. Read more about these analysis tools and other single-prison cell articles on the 10x Genomics Blog where we highlight of import research contributions, provide tips and tricks, and keep y'all up-to-appointment on all things single cell!
If you have any other questions or concerns about getting started with the Chromium Unmarried Jail cell 3' Solution, experience free to get out a comment here or contact u.s. directly. Whether it exist tracking brain organoid development, studying os marrow transplantation in patients with acute myeloid leukemia, or unraveling a novel pathway for intestinal stem cell cocky-renewal, the number of Chromium Unmarried Cell publications are growing daily, and we look forward to hearing about all of your astonishing enquiry!
Source: https://www.10xgenomics.com/blog/single-cell-rna-seq-an-introductory-overview-and-tools-for-getting-started
Posted by: hallettpasper.blogspot.com

0 Response to "how to find dropout rate in 10x genomics data"
Post a Comment